Replication stress response
DNA replication is essential for cell survival. To ensure that genetic information is preserved during proliferation, cells must duplicate of their entire genomic content before cell division. This is a challenging task, not least due to the fact that DNA is highly susceptible to damage. Some types of DNA lesions interfere with proper progression and completion of the replication process, thereby imposing so called “replication stress”. Importantly, replication stress has been recognised as one of the major factors that contributes to genomic instability associated with the development of human cancers.
Fortunately, cells have evolved mechanisms that protect them from a wide range of genotoxic insults. Strategies to resolve replication blocks are incredibly complex, and involve an intricate network of protein factors. We are interested in identifying novel players in this process, as well as exploring unknown functions of previously identified replication stress response proteins.
We have identified ZRANB3 as a novel factor implicated in the replication stress response (Weston et al, Genes Dev. 2012). ZRANB3 localizes to the sites of ongoing DNA replication and interacts with PCNA- a central scaffold protein that coordinates DNA replication and acts as a processivity factor for eukaryotic DNA polymerases. ZRANB3 is recruited to DNA breaks and stressed replication forks, and its deficiency leads to an increased sensitivity to certain DNA damaging agents. Structurally, ZRANB3 belongs to the SNF2 family of ATPases and contains several identifiable domains and motifs, which provide specificity for a number of defined substrates (including PCNA and K63-polyubiquitin chains).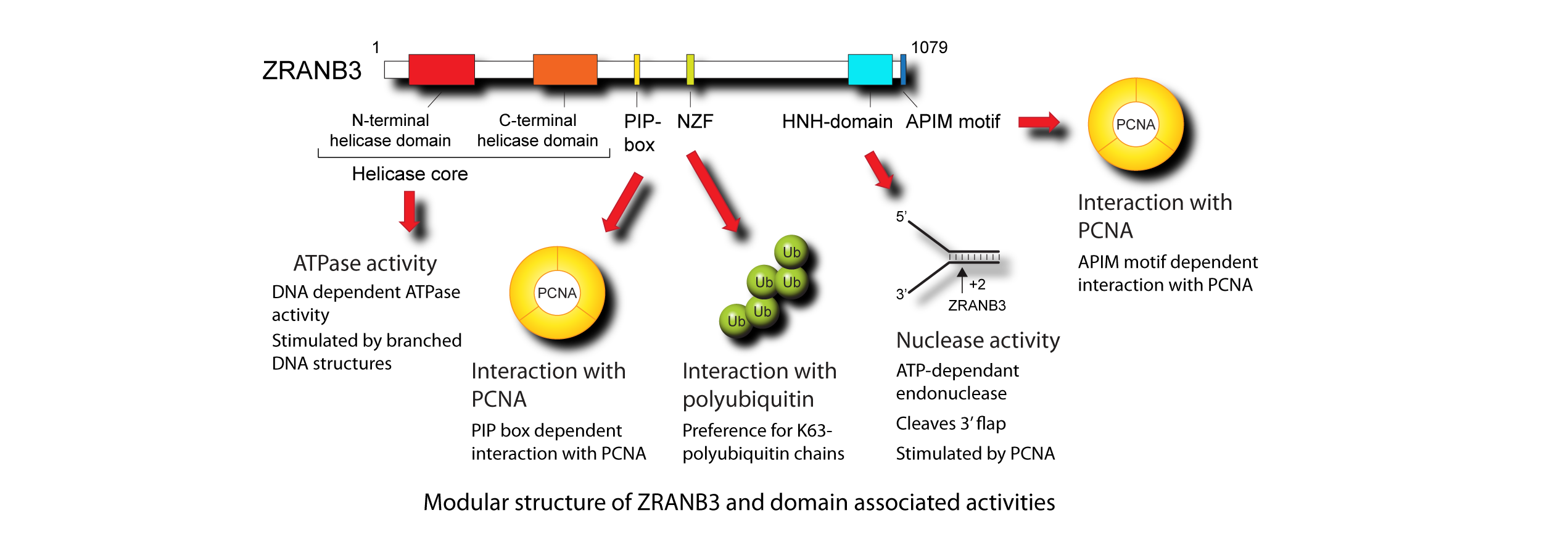
Importantly, we have shown that ZRANB3 acts as a highly unusual structure-specific endonuclease and cleaves its DNA substrate in an ATP-dependent manner. The endonuclease activity of ZRANB3 is supported by the C-terminal HNH domain, structurally related to the Holliday junction resolvase T4 endonuclease VII (Sebesta et al, Nat Commun. 2017). Interestingly, PCNA acts as a key regulator of ZRANB3 function: it both stimulates ZRANB3 endonuclease activity, and recruits ZRANB3 to stalled replication forks.
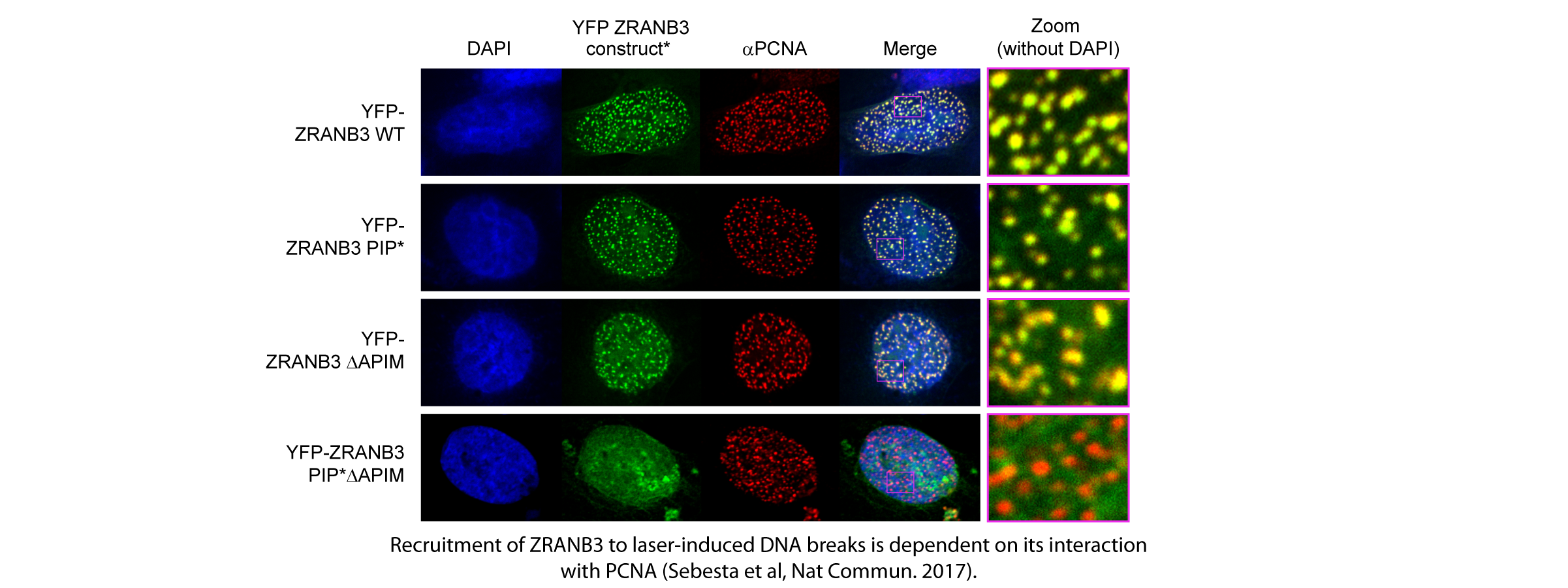
Our recent structural studies also provide insight into the molecular basis of ZRANB3 interactions with PCNA, mediated through two motifs in ZRANB3, the PIP box (Q-x-x-[VILM]-x-x-[FY]-[FY]) and the APIM motif ([KR]-[FYW]-[LIVA]-[LIVA]-[KR]). Our data provide the first structural insight into the binding of an APIM motif to PCNA and reveal unexpected similarities between the PIP box and the APIM motif: despite the differences in amino acid sequence, the two motifs share similar topology and bind to the same molecular surface of the PCNA ring.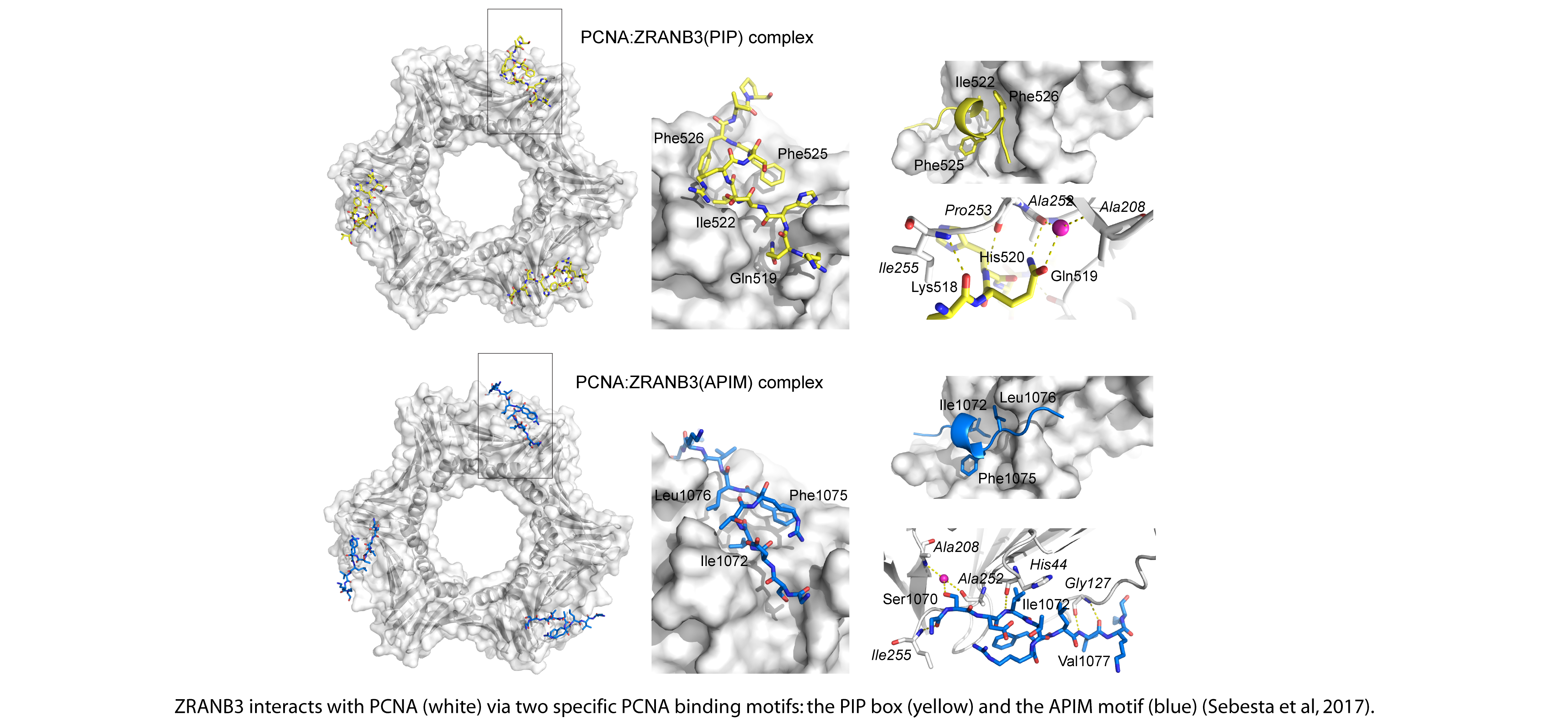
We are also interested in exploring the pathobiological importance of the ZRANB3 function. Multiple ZRANB3 variants have been identified in endometrial carcinoma, a malignancy characterised by marked genomic instability. Our structural and biochemical analyses enabled us to assess the functional relevance of cancer associated mutations of ZRANB3. Our data suggest a high frequency of loss-of-function mutations in ZRANB3 that may impact on its role in the replication stress response, and consequently on genomic stability.